About Us
Who we are and what we do?
Krasileva lab is an interdisciplinary group of people with a common interest in genomics and host-microbe interactions. Our vision is to combine the latest technologies (such as sequencing, synthetic biology, genome editing) with research to answer fundamental questions about genome evolution, eukaryotic innate immune systems.
Big Biological Question
The innate immune system is ancient and shared across kingdoms. Many organisms successfully utilize innate immune systems in the absence of an adaptive immunity to defend themselves against rapidly evolving pathogens: viruses, bacteria, fungi, and insects. Our laboratory works with several host-pathogen systems (mostly plants, bacteria and fungi) to understand biodiversity of host defenses, biodiversity of pathogens as well as develop strategies for engineering innate immunity
The major research areas and open questions in the lab are:
- Biodiversity of innate immune systems. How do hosts stay ahead of the pathogen?
- Mutagenesis, and synthetic biology. How can we engineer defense responses?
- Pathogen evolution and biodiversity. How do the pathogens strike back?
How do organisms with innate immunity alone (no adaptive immunity) recognize rapidly evolving pathogens?
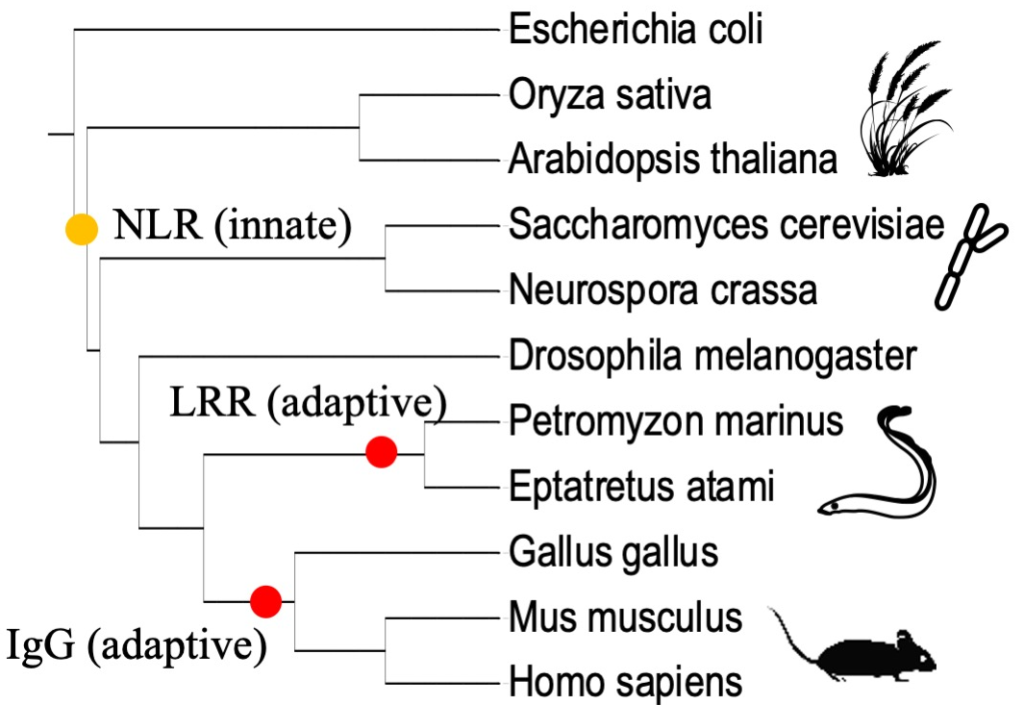 Evolution of NLR immune receptors: The main discoveries in this area include realization that similar protein families function in innate immunity across kingdoms. Our lab leads expertise in NLR evolution. We look at three main types of natural variation: 1) copy number variation, 2) structural variation, 3) allelic variation. The NLR protein family expansions and contractions and the evolution of downstream signaling pathways have been extensively investigated by PhD student Erin Baggs, now a postdoctoral fellow. (Baggs, Dagdas and Krasileva, 2017, Baggs et al, 2020, Michael et al, 2020). Previously, we also described a new type of diversity in NLRs – fusions to other plant proteins that serve as ‘baits’ for the pathogen (Sarris et al 2016, Bailey et al, 2018). Such NLRs with integrated domains (NLR-IDs) highlight plant proteins targeted by the pathogens and help to identify new sources of disease resistance. Our most recent work examined intra-species receptor diversity and identified hypervariable NLRs (hvNLRs) (Prigozhin and Krasileva, 2021). Using sequence information together with structural modeling, we are now able to classify immune receptors of an organism into functional groups as well as predict ligand binding sites. A mathematics graduate student Boyan Xu is currently looking at repeat evolution within NLRs. A new project led by a graduate student, Chandler Sutherland, aims to uncover precise molecular enzymes that control the diversity generation mechanism in the innate immune system, drawing parallels with how diversity is generated in adaptive immunity.
Evolution of NLR immune receptors: The main discoveries in this area include realization that similar protein families function in innate immunity across kingdoms. Our lab leads expertise in NLR evolution. We look at three main types of natural variation: 1) copy number variation, 2) structural variation, 3) allelic variation. The NLR protein family expansions and contractions and the evolution of downstream signaling pathways have been extensively investigated by PhD student Erin Baggs, now a postdoctoral fellow. (Baggs, Dagdas and Krasileva, 2017, Baggs et al, 2020, Michael et al, 2020). Previously, we also described a new type of diversity in NLRs – fusions to other plant proteins that serve as ‘baits’ for the pathogen (Sarris et al 2016, Bailey et al, 2018). Such NLRs with integrated domains (NLR-IDs) highlight plant proteins targeted by the pathogens and help to identify new sources of disease resistance. Our most recent work examined intra-species receptor diversity and identified hypervariable NLRs (hvNLRs) (Prigozhin and Krasileva, 2021). Using sequence information together with structural modeling, we are now able to classify immune receptors of an organism into functional groups as well as predict ligand binding sites. A mathematics graduate student Boyan Xu is currently looking at repeat evolution within NLRs. A new project led by a graduate student, Chandler Sutherland, aims to uncover precise molecular enzymes that control the diversity generation mechanism in the innate immune system, drawing parallels with how diversity is generated in adaptive immunity.
Do fungi also have innate immune responses? Fungi also have NLR receptors, yet their role in fungal immunity has not been tested. Although fungi are ubiquitous and often are known for their detrimental or beneficial interactions with other organisms, surprisingly little is known about fungal immune system. Knowing that most eukaryotic organisms have NLR-mediated innate immunity, PhD student Grace Stark decided to investigate what is the basis of innate immunity in fungi. She is working with Neurospora crassa - Pseudomonas system. Strikingly, Pseudomonas is able to colonize and grow along fungal hyphae.
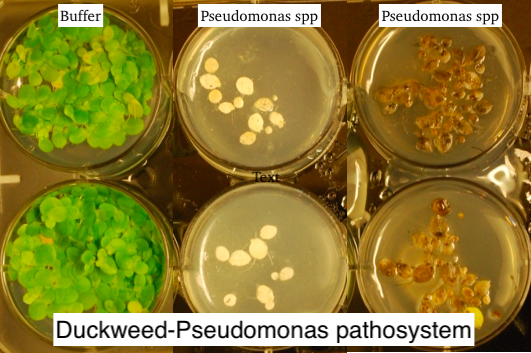
What about organisms that do not have NLRs? Although NLR receptors are present in most eukaryotes (including humans), they have been lost in some species, such as yeast. Erin Baggs discovered several independent plant lineages that have lost most NLRs as well as an EDS1/PAD4 innate immune signaling pathway. How do these plants cope with infections? Erin has set up a new duckweed - Pseudomonaspathosystem (Baggs et al 2022) and is addressing how duckweed that is surrounded by pathogens in its natural pond environment is capable of defending itself while missing several components of the canonical plant immune system. She is examining the contribution of antimicrobial proteins expanded in duckweeds as well as recruitment of other bacteria by duckweed to defend itself against pathogens.
Can we find new ways to boost immunity?
Engineering plant immunity using mutagenesis and synthetic biology: We are rapidly transferring our knowledge of NLR evolution into novel strategies of engineering plant immune receptors (Tamborski and Krasileva, 2020). As one strategy, we use a rational design of synthetic plant immune receptors by a gene fusion process. The second strategy builds on our discovery of hypermutating NLRs and our ability to computationally identify hypervariable ligand binding sites. We aim to deploy both principles to generate synthetic plant immune receptors with new ‘baits’ against pathogen molecules. We have been successfully able to rationally change receptor specificity and create a synthetic receptor by modifying as few as 12 amino acids (Tamborski et al 2022). With the advancement of DNA synthesis and protein engineering, we are also embarking on new ways to screen for new NLR specificites using high-throughput screening in yeast led by a postdoc Lorena Parra. Postdoc in the lab, Wei Wei, is continuing to explore synthetic biology and rational design of NLR circuits.
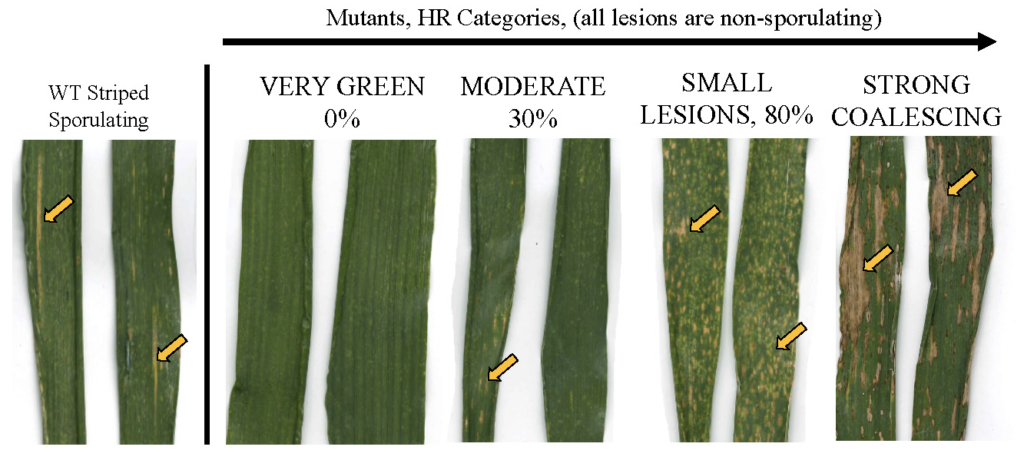 To complement our approaches with unbiased forward screens, we have conducted a field screen of EMS-mutagenized wheat and identified over thirty mutants with dominant enhanced disease resistance against a devastating obligate biotrophic fungal pathogen called wheat yellow rust (Puccinia striiformis f. sp. tritici) and having diverse phenotypes showing extensive lesion formation or nearly none. These lines have also been exome-captured, providing a public reverse genetics resource as well. We are prioritizing the identification of causal mutations responsible for resistance as well as to characterize physiological pathways providing wheat with an immune boost, for eventual promoter mining. We leverage the cold-inducible, autoimmune nature of several of these mutants to work with them under growth chamber conditions. We are also investigating possible biotrophic vs necrotrophic tradeoffs in susceptibility by screening the same mutants against Septoria, which infects dead tissue as found in mutants that make leaf lesions to restrict rust growth. A lab manager China Shaw and a postdoc Rakesh Kumar are involved in this project. Two mutant lines of particular interest maintain very green leaves in the presence of both pathogens and maintain a thousand kernel weight at least as high as wildtype, in the presence of yellow rust.
To complement our approaches with unbiased forward screens, we have conducted a field screen of EMS-mutagenized wheat and identified over thirty mutants with dominant enhanced disease resistance against a devastating obligate biotrophic fungal pathogen called wheat yellow rust (Puccinia striiformis f. sp. tritici) and having diverse phenotypes showing extensive lesion formation or nearly none. These lines have also been exome-captured, providing a public reverse genetics resource as well. We are prioritizing the identification of causal mutations responsible for resistance as well as to characterize physiological pathways providing wheat with an immune boost, for eventual promoter mining. We leverage the cold-inducible, autoimmune nature of several of these mutants to work with them under growth chamber conditions. We are also investigating possible biotrophic vs necrotrophic tradeoffs in susceptibility by screening the same mutants against Septoria, which infects dead tissue as found in mutants that make leaf lesions to restrict rust growth. A lab manager China Shaw and a postdoc Rakesh Kumar are involved in this project. Two mutant lines of particular interest maintain very green leaves in the presence of both pathogens and maintain a thousand kernel weight at least as high as wildtype, in the presence of yellow rust.
How do pathogens evolve virulence, escape recognition and change hosts?
Fungal pathogens rapidly adapt to new environments, hosts and lifestyles. In the pathogen genomes, the signature of the adaption appears as a two-speed genome: slowly evolving regions carry housekeeping genes, and rapidly evolving regions contain virulence genes that cause disease. The rapid evolution of fungal pathogens is also reflected in their virulence proteins named effectors: many effectors that display no sequence similarity structurally look alike, possibly due to divergent evolution. Our lab uses genomics and computational approaches to understand pathogen evolution and the diversity of the pathogens.
Structural variation in blast pathogens: The rapidly evolving regions of the fungal phytopathogen genomes are rich in repeats and transposons. Extrachromosomal circular DNAs (eccDNAs) are thought to originate from such genomic regions and help the pathogens to respond to stress by dramatically increasing the gene copy number. EccDNAs were recently isolated from the rice blast fungus Magnaporthe oryzae and their role in response to stress by the pathogen has been investigated by Pierre Joubert (Joubert and Krasileva, 2021). Pierre is now continuing with comparative genomics analyses across Magnaporthe to investigate and model structural variation to answer a question how a fungal pathogen can rapidly gain and lose genes, including potential virulence factors.
The diversity of transposable elements in blast pathogens: Transposable elements (TEs) may also play a role in two-speed genome evolution by generating genomic diversity. The evolution of lineage-specific TE expansions in M. oryzae is being investigated by Anne Nakamoto, with the goal of understanding how TEs might play a role in the pathogen’s adaptation to a wide host range.
Elucidating structural similarity between fungal effectors with AlphaFold: Identifying sequence-unrelated structurally similar (SUSS) effectors has been important to the community, due to the SUSS effectors’ essential roles in the plant-pathogen interaction. Graduate student in the lab, Kyungyong Seong, developed novel approaches using computational structural genomics to identify new and existing classes of SUSS effectors (Seong and Krasileva, 2021). Our most recent study in this area expands to diverse phytopathogens across the fungal kingdom to highlight divergent evolution of fungal effectors commonly observed in different species (Seong and Krasileva, 2022).
Recent talks by the lab
Ksenia - Catching the right mutation - enhanced disease resistance mutants in wheat protect against stripe rust and other pathogens, America Phytopathological Society, 2021
Ksenia - The highly variable plant immune receptors and how to catch them, iCAR, 2021
Our research is funded by