About Us
Who we are and what we do
We are an interdisciplinary group of people with shared interests in comparative immunology, biology of genomes and host-microbe interactions. Our vision is to combine fundamental research with modern computational biology to understand biodiversity of host defenses, pathogens as well as to develop strategies for predictive modeling and engineering innate immunity
The major research areas and open questions in the lab are:
- How do hosts stay ahead of the pathogen?
- How can we engineer defense responses?
- How do the pathogens strike back?
Big Biological Question: How do organisms with innate immunity alone (no adaptive immunity) recognize rapidly evolving pathogens?
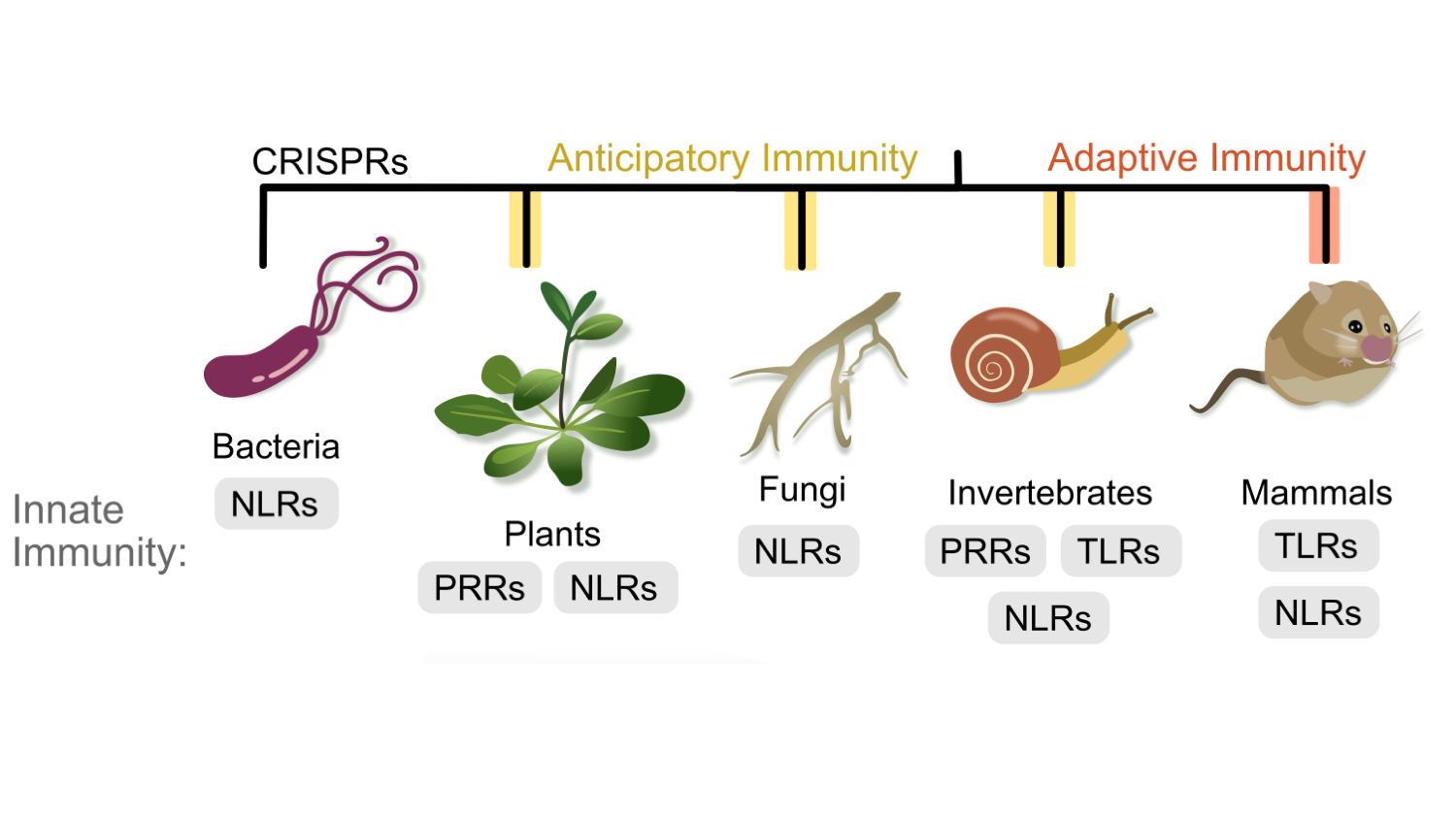
Evolution of NLR immune receptors: All organisms have an innate immune system, including bacteria, fungi, plants, and animals. Moreover, similar protein receptors function in innate immunity across kingdoms. Our lab leads expertise in comparative immunology and innate immune receptor evolution. We also investigate how understanding diversity of innate immune systems can drive new biotechnological approaches.
Historically, we have been looking at plants (check out our most recent review). More recently, we added fungi and started looking at fungal immune systems. We are also open to discussion and potential collaborations on other branches of life.
Recent talks by the lab
Ksenia - Catching the right mutation - enhanced disease resistance mutants in wheat protect against stripe rust and other pathogens, America Phytopathological Society, 2021
Ksenia - The highly variable plant immune receptors and how to catch them, iCAR, 2021
- From Natural Diversity to Precise Modifications of Plant Immune Receptors
Plants are natural engineers of new pathogen recognition specificities. We have utilized our knowledge about receptor diversity towards understanding and engineering of intracellular immune receptors (NLRs), with publications focusing on structural annotation, functional resurrection of activity against pathogen variants that escaped detection. Our computational contributions span from deep learning models to pan-genomic feature extraction and engineering efforts include precise prediction of 2-3 amino acid changes needed to restore pathogen recognition, providing an avenue for future genome editing of native alleles in the genomes, an approach that we call “immunotherapy for plants”.
- Highly Variable Receptors have Distinct Genomic Features: We mapped high allelic diversity of Arabidopsis NLRs (Sutherland et al, EMBO Reports, 2024) and linked it to distinct genomic signatures, providing insight into evolutionary constraints and opportunities for synthetic biology. The analyses also hint at possibility of the natural ‘mutators’ that act on immune loci, an active research area that we are pursuing now with newest sequencing technologies. We also continued our work on the annotation of structural variations and presence/absence variation in immune receptor loci, especially in maize (Prigozhin et al. MPMI, 2024) and wild tomato (Seong et al bioRxiv 2022). We released chromosome-level assemblies for wheat (Seong et al Zenodo, 2023) and wild tomato (Seong et al, bioRxiv 2022), uncovering structural variation important for NLR function. These datasets support global efforts to harness wild relatives for crop improvement. These studies combine long-read genomics with in-house developed tools and were made fully open-source on GitHub and Zenodo.
- Immune Receptor as a Slinky: Mathematical Structure-aware Annotation of Protein Repeats: We introduced a new mathematical method for structure-guided annotation of ligand binding repeats in solenoid proteins, such as plant immune receptors (Xu et al PLoS Comp Biol, 2024), which we now use to curate immune receptor databases and facilitate precision engineering. This is a truly interdisciplinary work led by talented mathematics student Boyan Xu who decided to shift his PhD work towards biology and gained Designed Emphasis in Computational Biology in our lab.
- Resurrecting Disease Resistance with Precise Modifications: In bioRxiv(Seong et al, 2024), a graduate student in our lab Kyungyong Seong developed the successful redesign of the wheat NLR receptor to recognize an escape variant of the pathogen-encoded effector molecule, overcoming a major hurdle in wheat disease resistance. This work demonstrates rational engineering based on structural predictions, ability to quickly resurrect a defeated immune receptor with minimal modifications and expand the toolkit for sustainable resistance breeding. Separately, a postdoc Wei Wei applied synthetic biology approach to engineer pathogen-inducible transcription of selected immune genes in tomato (Wei et al bioRxiv 2024). In another study in Molecular Breeding (Lunde et al, 2025) characterizes durum wheat mutants with enhanced resistance to stripe rust and differential responses to other pathogens pinpointing induced changes in different pathways that lead to gain of resistance, the study includes multi-year field studies and collaborative testing against multiple pathogens. This work informs trait stacking strategies and underpins ongoing breeding collaborations.
- Predicting Immunogenicity of Receptor-Ligand Interactions via Deep Learning: In our most recent work, we developed mamp-ml, a machine learning fine-tuned language model pipeline for predicting the immunogenicity of microbial epitopes in plants (Stevens et al bioRxiv, 2025). This is the first such platform tailored to plant immunity that is gaining strong recognition across several fields and was selected for oral presentation after full conference peer review at Machine Learning in Computational Biology 2025.
The field is rapidly evolving and our laboratory continues to maintain its expert status by blending emerging computational technologies with functional analyses. We have been invited to contribute a Plant Cell review (Sutherland et al 2025) which synthesizes findings across Arabidopsis and crops, highlighting how natural variation at DNA, RNA, and protein levels can inform receptor design. One of the next challenges is translational research – deployment of these findings in industrial scale crop production. During 2024-2025, Ksenia has served as guest editor of the Molecular Plant Microbe Interactions Special Issue: “Fine Grain: Molecular, Cellular, and Genomic Details of Cereal Crop Diseases” that organized research community to present the most recent research in the area and provided tangible solutions going towards disease resistance deployment and she will serve on the Molecular Plant Microbe Interactions editorial board starting January 2026.
- Evolution in Overdrive: Fungal Pathogen Genomes and Rapidly Evolving Secreted Proteins
Our lab continues to be a leader in the field of fungal pathogen genomics and understanding of secreted proteins translocated by pathogens inside their hosts (effectors). The research led in our laboratory as well as collaborations do not focus on a single pathogen, rather we use modern computational tools and comparative approaches to understand 1) how does a fungus become a pathogen or switch its hosts and 2) how can we computationally prioritize pathogen-encoded factors for functional studies.
- Structural Variation in Magnaporthe Genomes Encode Host Specificity: We published a series of papers on Magnaporthe oryzae, the blast fungus of wheat and rice, also considered the top fungal threat to crop production worldwide (Seong and Krasileva, Nature Microbiology 2021; Joubert, and Krasileva, BMC Biology 2023; Nakamoto, Joubert, and Krasileva Genome Biol Evol 2023; Joubert and Krasileva, Genetics, 2024)
- In one seminal study led by a post-bac student, Anne Nakamoto (now a graduate student at UC Santa Cruz), we investigated the role of transposable elements (Nakamoto, Joubert, and Krasileva Genome Biol Evol, 2023) in evolution of new fungal strains and host specificity. In another study, a graduate student in the lab Pierre Joubert not only documented differences in genomic structural variation in determining pathotype-specific gene content (Joubert and Krasileva, Genetics, 2024) but also constructed random forest based predictive models.
Our previous key study on effector evolution across 24 fungal species using comparative computational structural biology (Seong and Krasileva, Nature Microbiology 2023) established not only established us as experts in the field of comparative structural genomics, but also led to multiple collaborations.
- Novel conserved effectors in Smut Fungi: In collaboration with international partners, we identified novel conserved effectors in Ustilago maydisthat contribute to virulence (MPMI, 2024). This work is foundational for comparative effector biology in smut fungi.
- Zymoseptoria Effectors Suppressing Host Immunity: A collaborative study (Mol Plant Pathol, 2024) uncovered an array of triticieffectors that suppress host immunity. This provides mechanistic insight into how complex pathogen populations evade detection.
These studies continue to uncover diverse strategies of fungal pathogenesis, inform disease resistance deployment and contribute to comparative genomics of high-impact fungal pathogens.
- Discovery of Non-canonical Immune Systems
We pioneered new experimental systems, including establishing a fully genetically tractable system for studying filamentous fungus – bacterial interactions.
- Fungal Immunity in a Novel Model: In a study led by graduate student Frances Grace Stark, we demonstrated that Neurospora crassacan mount innate immune responses to Pseudomonas syringae (Stark et al bioRxiv, 2025) and functionally characterized several fungal genes that maintain fungal fitness upon bacterial interactions. This work establishes a genetically tractable fungal model for innate immunity and allows us to test hypotheses about conserved and innovative strategies fungi deploy for recognition and response to other microbes.
- Beneficial Pseudomonas Strains: Another non-canonical immune system can be found in duckweed, a clonally propagating invasive aquatic plant species which lacks most of the well described immune genes and pathways characteristic of land plants. Our duckweed microbiome study (Baggs, Stark et al bioRxiv, 2022) found native Pseudomonas isolates that suppress disease from pathogenic strains, highlighting underexplored mechanisms of microbe-microbe competition.
Collaborations within our department: We actively collaborate within our department, as reflected in two recent studies: one with Yangnan Gu’s laboratory (Tang Y et al Nature Plants 2024) where we provided our expertise on computational structural biology and the second with Patrick Shih’s laboratory (Alamos et al Nature Plants 2025) where we served as experts and provided molecular assessment of plant innate immune response induction.
Open Science and approach to research mentorship: All code developed by our lab is openly available, and we have deposited over a dozen new datasets with DOIs. Preprints are posted on bioRxiv ahead of submission to foster transparency and reproducibility. Graduate students and postdocs are lead authors on nearly all papers, reflecting the lab’s strong culture of mentorship and collaborative research.
This research program and outputs reflect our lab’s central role in shaping the future of plant immune engineering, pathogen genomics, and integrative computational biology. They support UC Berkeley’s mission of high-impact interdisciplinary research and scientific leadership.
Our research is funded by


![]()